RKeOps
State of the R
24-28/08/2020
Motivation: scalable kernel operations
Sources:
- Presentation of Ghislain Durif at useR! 2020
- Documentation of KeOps
KeOps stands for “Kernel Operations”. RKeOps is an R package interfacing the KeOps library.
Kernels are widely used in Statistics and Learning:
- Kernel density estimation
- Classification/Regression: SVM, K-NN, etc…
- Kernel embeddings to compare distributions
- Interpolation and Kriging
- Optimal Transport
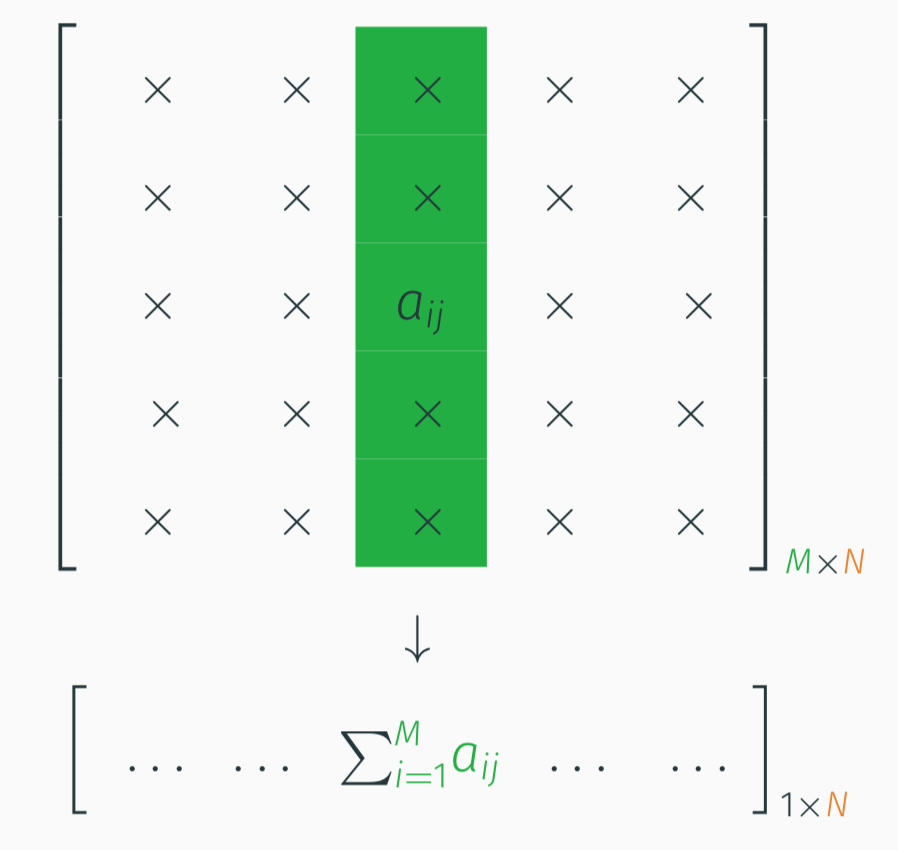
The main motivation behind KeOps is the need to compute fast and scalable kernel operations such as Gaussian convolutions (RBF kernel products). For very large values of \(M\)and \(N\), given :
- a target point cloud \((x_i)_{i=1}^M \in \mathbb R^{M \times D}\),
- a source point cloud \((y_j)_{j=1}^N \in \mathbb R^{N \times D}\),
- a signal \((b_j)_{j=1}^N \in \mathbb R^{N}\) attached to the \(y_j\)’s,
KeOps allows you to compute efficiently the array \((a_i)_{i=1}^M \in \mathbb R^{M}\) given by
\[ a_i = \sum_j K(x_i,y_j) b_j, \qquad i=1,\cdots,M,\] where \(K\) is e.g. the Gaussian Kernel: \(K(x,y) = \exp\left(-\Vert x_i - y_j \Vert^2/(2\sigma^2) \right)\).
These types of operations involve reductions of very large arrays e.g. row-wise or column-wise matrix sums:
Automatic differentiation
Thanks to KeOps’ automatic differentiation module, you can also get access to the gradient of the \(a_i\)’s with respect to the \(x_i\)’s:
\[ a_i' = \sum_{j=1}^N \partial_x K(x_i,y_j) b_j, \qquad i=1,\cdots,M,\]
without having to code the formula \(\partial_x K(x_i,y_j) = -\tfrac{1}{\sigma^2}(x_i - y_j) \exp(-\|x_i - y_j\|^2 / 2 \sigma^2)\)!
One-slide summary

Installation
Reference: Installation page
The package is only available from UNIX-type platforms
Installation requires cmake.
To install cmake on a Mac one can use:
brew install cmakeThen the rkeops package can be installed from CRAN:
if (!require("rkeops")) {
install.packages("rkeops")
}Getting started
library("rkeops")##
## You are using rkeops version 1.4.1.1RKeOps allows to define and compile new operators that run computations on GPU.
# implementation of a convolution with a Gaussian kernel
formula = "Sum_Reduction(Exp(-s * SqNorm2(x - y)) * b, 0)"
# input arguments
args = c("x = Vi(3)", # vector indexed by i (of dim 3)
"y = Vj(3)", # vector indexed by j (of dim 3)
"b = Vj(6)", # vector indexed by j (of dim 6)
"s = Pm(1)") # parameter (scalar)Then we compile the formula:
# compilation
op <- keops_kernel(formula, args)## Compiling headersb72ec1124da8340d0118a99af in /usr/share/miniconda/envs/finistR2020/lib/R/library/rkeops:
## formula: Sum_Reduction(Exp(-s * SqNorm2(x - y)) * b, 0)
## aliases: decltype(Vi(0,3)) x ;decltype(Vj(1,3)) y ;decltype(Vj(2,6)) b ;decltype(Pm(3,1)) s ;
## dtype : float
## ...# data and parameter values
nx <- 100
ny <- 150
X <- matrix(runif(nx*3), nrow=nx) # matrix 100 x 3
Y <- matrix(runif(ny*3), nrow=ny) # matrix 150 x 3
B <- matrix(runif(ny*6), nrow=ny) # matrix 150 x 6
s <- 0.2If your machine is GPU friendly gpu computation can be turned on, otherwise CPU should be used
# use_gpu()
use_cpu()Finally we can perform the desired kernel operation:
# computation (order of the input arguments should be similar to `args`)
res <- op(list(X, Y, B, s))
str(res)## num [1:100, 1:6] 58.9 58.6 60.4 61 61.9 ...The author of the package is very responsive: he has fixed a Mac-specific compiling issue I had when trying to run this vignette.
Session information
sessionInfo()## R version 3.6.3 (2020-02-29)
## Platform: x86_64-conda_cos6-linux-gnu (64-bit)
## Running under: Ubuntu 18.04.5 LTS
##
## Matrix products: default
## BLAS/LAPACK: /usr/share/miniconda/envs/finistR2020/lib/libopenblasp-r0.3.10.so
##
## locale:
## [1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C
## [3] LC_TIME=C.UTF-8 LC_COLLATE=C.UTF-8
## [5] LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
## [7] LC_PAPER=C.UTF-8 LC_NAME=C.UTF-8
## [9] LC_ADDRESS=C.UTF-8 LC_TELEPHONE=C.UTF-8
## [11] LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C.UTF-8
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] rkeops_1.4.1.1 optimLibR_0.1.0 slider_0.1.5
## [4] fable_0.2.1 fabletools_0.2.1 tsibble_0.9.2
## [7] vip_0.2.2 skimr_2.1.2 yardstick_0.0.7
## [10] workflows_0.1.3 tune_0.1.1 rsample_0.0.7
## [13] recipes_0.1.13 parsnip_0.1.3 modeldata_0.0.2
## [16] infer_0.5.3 dials_0.0.8 scales_1.1.1
## [19] broom_0.7.0 tidymodels_0.1.1 MASS_7.3-52
## [22] mgcv_1.8-33 nlme_3.1-149 fda_5.1.5.1
## [25] Matrix_1.2-18 deSolve_1.28 GGally_2.0.0
## [28] ggdist_2.2.0 distributional_0.2.0 DT_0.15
## [31] forcats_0.5.0 stringr_1.4.0 dplyr_1.0.2
## [34] purrr_0.3.4 readr_1.3.1 tidyr_1.1.2
## [37] tibble_3.0.3 ggplot2_3.3.2 tidyverse_1.3.0
##
## loaded via a namespace (and not attached):
## [1] readxl_1.3.1 backports_1.1.9 RcppEigen_0.3.3.7.0
## [4] plyr_1.8.6 repr_1.1.0 lazyeval_0.2.2
## [7] splines_3.6.3 crosstalk_1.1.0.1 listenv_0.8.0
## [10] digest_0.6.25 JuliaCall_0.17.1.9000 foreach_1.5.0
## [13] htmltools_0.5.0.9000 fansi_0.4.1 magrittr_1.5
## [16] globals_0.12.5 modelr_0.1.8 gower_0.2.2
## [19] askpass_1.1 hardhat_0.1.4 anytime_0.3.9
## [22] prettyunits_1.1.1 jpeg_0.1-8.1 colorspace_1.4-1
## [25] blob_1.2.1 rvest_0.3.6 warp_0.1.0
## [28] haven_2.3.1 xfun_0.16 crayon_1.3.4
## [31] jsonlite_1.7.1 progressr_0.6.0 survival_3.2-3
## [34] iterators_1.0.12 glue_1.4.2 gtable_0.3.0
## [37] ipred_0.9-9 DBI_1.1.0 Rcpp_1.0.5
## [40] viridisLite_0.3.0 GPfit_1.0-8 lava_1.6.7
## [43] prodlim_2019.11.13 htmlwidgets_1.5.1 httr_1.4.2
## [46] RColorBrewer_1.1-2 ellipsis_0.3.1 pkgconfig_2.0.3
## [49] reshape_0.8.8 farver_2.0.3 nnet_7.3-14
## [52] dbplyr_1.4.4 utf8_1.1.4 tidyselect_1.1.0
## [55] labeling_0.3 rlang_0.4.7 DiceDesign_1.8-1
## [58] munsell_0.5.0 cellranger_1.1.0 tools_3.6.3
## [61] cli_2.0.2 generics_0.0.2 ranger_0.12.1
## [64] evaluate_0.14 yaml_2.2.1 knitr_1.29
## [67] fs_1.5.0 future_1.18.0 xml2_1.3.2
## [70] compiler_3.6.3 rstudioapi_0.11 plotly_4.9.2.1
## [73] png_0.1-7 reprex_0.3.0 lhs_1.0.2
## [76] stringi_1.4.6 highr_0.8 lattice_0.20-41
## [79] readODS_1.7.0 vctrs_0.3.4 diffeqr_1.0.0
## [82] nycflights13_1.0.1 pillar_1.4.6 lifecycle_0.2.0
## [85] furrr_0.1.0 data.table_1.12.8 R6_2.4.1
## [88] gridExtra_2.3 codetools_0.2-16 assertthat_0.2.1
## [91] openssl_1.4.2 withr_2.2.0 parallel_3.6.3
## [94] hms_0.5.3 grid_3.6.3 rpart_4.1-15
## [97] timeDate_3043.102 class_7.3-17 rmarkdown_2.3
## [100] pROC_1.16.2 lubridate_1.7.9 base64enc_0.1-3