Table of Contents
Introduction
library(tidyverse)
library(car)
library(broom)
library(flextable)
theme_set(theme_bw())Extrait des données
testScore | bodyLength | mountainRange | site |
7.359 | 179.606 | Bavarian | a |
51.536 | 203.515 | Emmental | a |
62.872 | 226.011 | Julian | c |
89.043 | 232.514 | Julian | c |
64.161 | 226.274 | Julian | c |
52.682 | 204.596 | Maritime | b |
82.854 | 195.131 | Sarntal | a |
75.246 | 203.173 | Sarntal | b |
80.293 | 201.469 | Sarntal | b |
21.169 | 183.459 | Southern | c |
Statistiques univariées
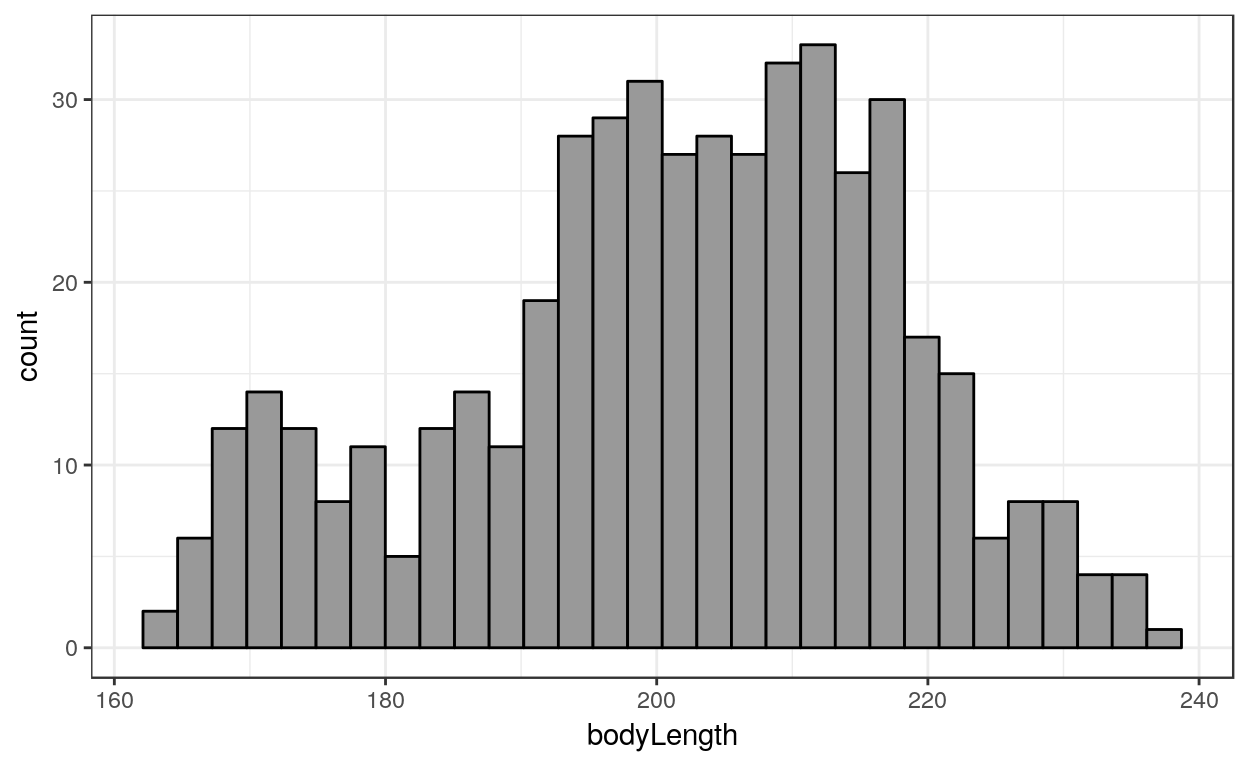
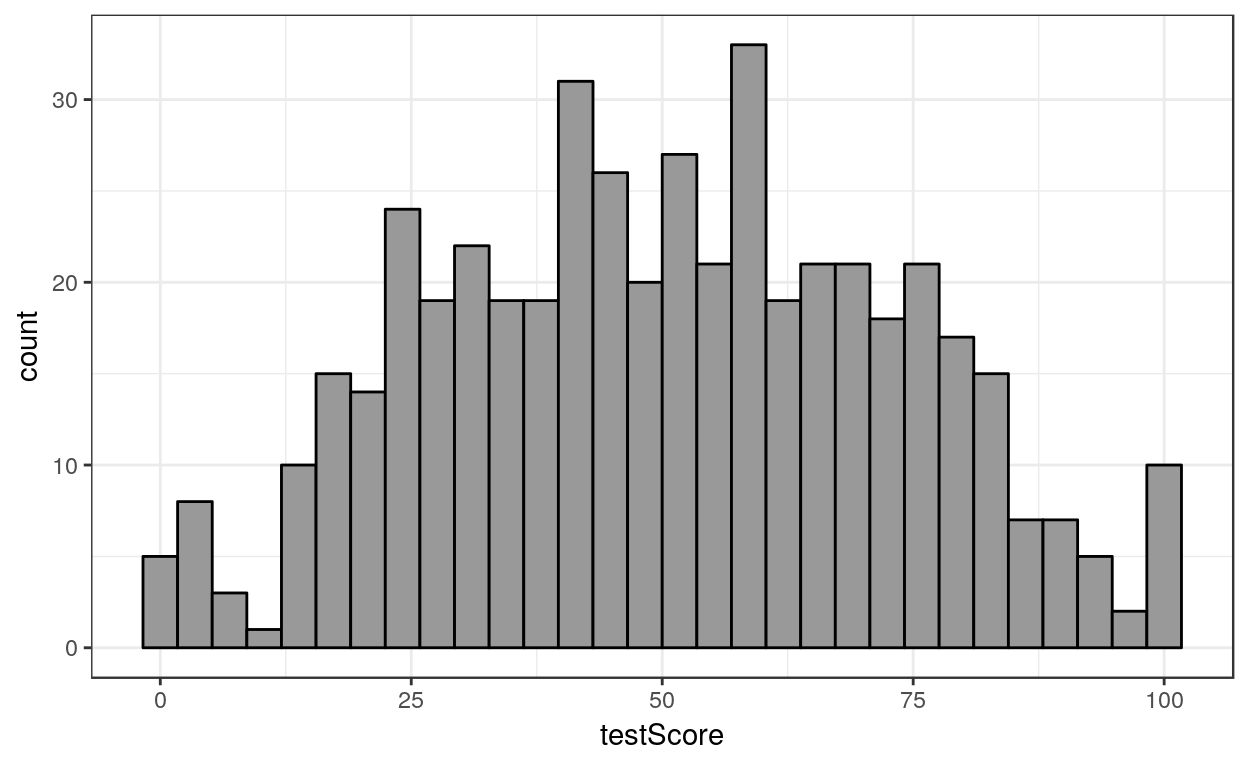
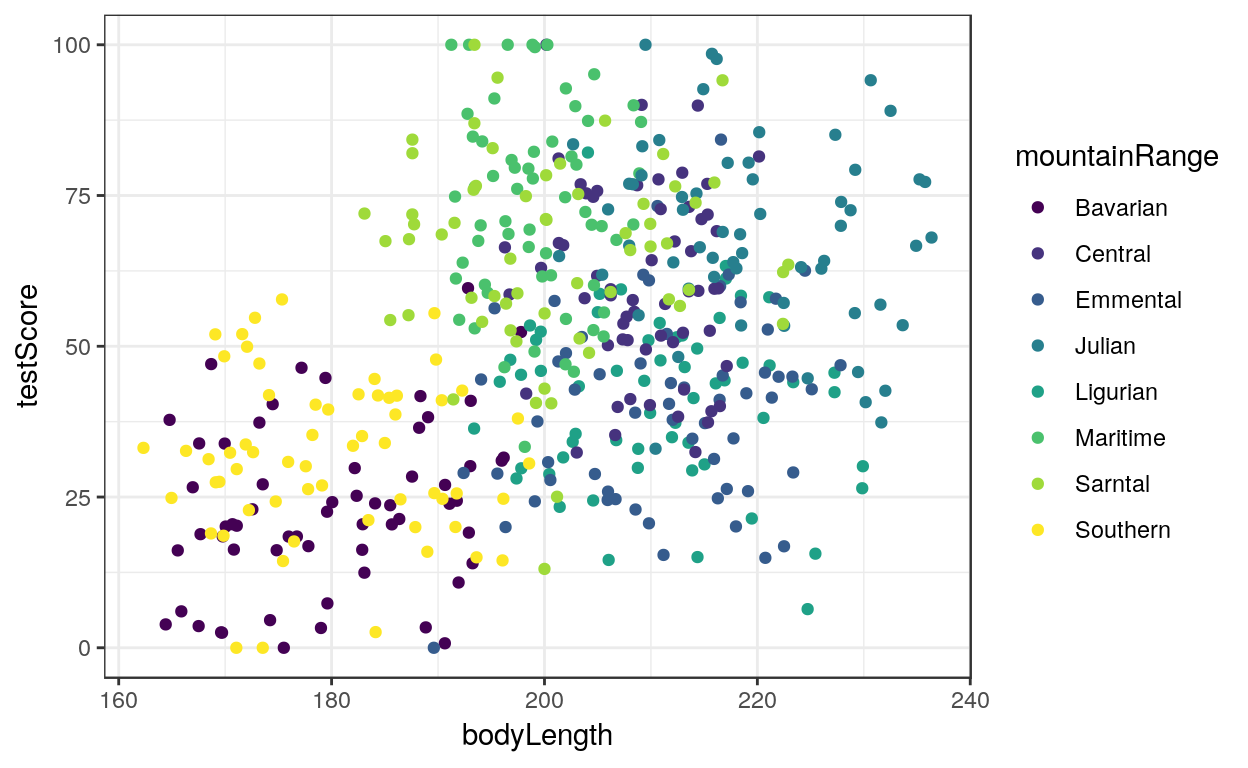
Analyse statistique
Modèle linéaire simple
model_simple <- lm(testScore ~ bodyLength, data = dragons)term | estimate | std.error | statistic | p.value |
(Intercept) | -61.318 | 12.067 | -5.081 | 0.000 |
bodyLength | 0.555 | 0.060 | 9.287 | 0.000 |
La longueur du dragon explique son intelligence.
ANCOVA
model_ancova <- lm(testScore ~ bodyLength + mountainRange, data = dragons)On regarde les coefficients de chaque variable via broom::tidy().
term | estimate | std.error | statistic | p.value |
(Intercept) | 20.831 | 14.472 | 1.439 | 0.151 |
bodyLength | 0.013 | 0.080 | 0.159 | 0.874 |
mountainRangeCentral | 36.583 | 3.599 | 10.164 | 0.000 |
mountainRangeEmmental | 16.209 | 3.697 | 4.385 | 0.000 |
mountainRangeJulian | 45.115 | 4.190 | 10.767 | 0.000 |
mountainRangeLigurian | 17.748 | 3.674 | 4.831 | 0.000 |
mountainRangeMaritime | 49.881 | 3.139 | 15.890 | 0.000 |
mountainRangeSarntal | 41.978 | 3.197 | 13.130 | 0.000 |
mountainRangeSouthern | 8.520 | 2.731 | 3.119 | 0.002 |
On fait une analyse de la variance de type II via car::Ancova().
term | sumsq | df | statistic | p.value |
bodyLength | 5.653 | 1.000 | 0.025 | 0.874 |
mountainRange | 109415.981 | 7.000 | 69.844 | 0.000 |
Residuals | 105407.810 | 471.000 |
La longueur du dragon n’est plus significative pour expliquer son intelligence si on prend en compte la région d’où il vient.
Modèle mixte
On utilise le package lme4 pour faire du modèle mixte et broom.mixed pour mettre en forme les résultats.
library(lme4)
library(broom.mixed)
model_mixed <- lmer(testScore ~ bodyLength + (1|mountainRange),
data = dragons, REML = TRUE)On regarde tidy(model_mixed) et glance(model_mixed).
effect | group | term | estimate | std.error | statistic |
fixed | (Intercept) | 43.709 | 17.135 | 2.551 | |
fixed | bodyLength | 0.033 | 0.079 | 0.422 | |
ran_pars | mountainRange | sd__(Intercept) | 18.430 | ||
ran_pars | Residual | sd__Observation | 14.960 |
sigma | logLik | AIC | BIC | REMLcrit | df.residual |
14.960 | -1995.601 | 3999.203 | 4015.898 | 3991.203 | 476 |
On va comparer ce modèle avec le modèle qui ne contient pas bodyLength.
model_mixed_null <- lmer(testScore ~ 1 + (1|mountainRange),
data = dragons, REML = TRUE)Puis on fait une anova de type I via anova().
term | df | AIC | BIC | logLik | deviance | statistic | Chi.Df | p.value |
model_mixed_null | 3.000 | 3999.685 | 4012.206 | -1996.843 | 3993.685 | |||
model_mixed | 4.000 | 4001.478 | 4018.173 | -1996.739 | 3993.478 | 0.207 | 1.000 | 0.649 |
La variable n’est pas bodyLength n’est pas significative.
Modèle mixte à effets imbriqués
model_nested <- lmer(testScore ~ bodyLength + (1|mountainRange) + (1|mountainRange:site),
data = dragons)effect | group | term | estimate | std.error | statistic |
fixed | (Intercept) | 40.067 | 21.864 | 1.833 | |
fixed | bodyLength | 0.051 | 0.104 | 0.494 | |
ran_pars | mountainRange:site | sd__(Intercept) | 4.805 | ||
ran_pars | mountainRange | sd__(Intercept) | 18.099 | ||
ran_pars | Residual | sd__Observation | 14.442 |
sigma | logLik | AIC | BIC | REMLcrit | df.residual |
14.442 | -1987.980 | 3985.961 | 4006.830 | 3975.961 | 475 |
model_nested_null <- lmer(testScore ~ 1 + (1|mountainRange) + (1|mountainRange:site),
data = dragons)On fait une anova de type un entre model_nested et model_null.
term | df | AIC | BIC | logLik | deviance | statistic | Chi.Df | p.value |
model_nested_null | 4.000 | 3987.054 | 4003.749 | -1989.527 | 3979.054 | |||
model_nested | 5.000 | 3988.767 | 4009.636 | -1989.384 | 3978.767 | 0.287 | 1.000 | 0.592 |
La modèle qui contient bodyLength n’est pas significativement différent du modèle qui ne le contient pas.
Notes
ML / REML
Par défaut, lmer() va optimiser REML et non pas ML. Cependant, lorsqu’on fait anova() entre deux modèles, ceux-ci sont recalculés pour être optimisés selon ML.
Prédiction
augment(model_nested, dragons)
# A tibble: 480 x 14
testScore bodyLength mountainRange site .fitted .resid .cooksd
<dbl> <dbl> <fct> <fct> <dbl> <dbl> <dbl>
1 16.1 166. Bavarian a 20.8 -4.62 2.38e-3
2 33.9 168. Bavarian a 20.9 13.0 1.82e-2
3 6.04 166. Bavarian a 20.8 -14.8 2.41e-2
4 18.8 168. Bavarian a 20.9 -2.04 4.47e-4
5 33.9 170. Bavarian a 21.0 12.9 1.73e-2
6 47.0 169. Bavarian a 20.9 26.1 7.21e-2
7 2.56 170. Bavarian a 21.0 -18.4 3.55e-2
8 3.88 164. Bavarian a 20.7 -16.8 3.24e-2
9 3.60 168. Bavarian a 20.9 -17.3 3.21e-2
10 7.36 180. Bavarian a 21.5 -14.1 2.16e-2
# … with 470 more rows, and 7 more variables: .fixed <dbl>,
# .mu <dbl>, .offset <dbl>, .sqrtXwt <dbl>, .sqrtrwt <dbl>,
# .weights <dbl>, .wtres <dbl>On a accès aux coefficients via les fonctions coefficients() pour les effets fixes et ranef() pour les effets aléatoires.
coefficients(summary(model_nested))
Estimate Std. Error t value
(Intercept) 40.06668990 21.8637329 1.8325640
bodyLength 0.05125931 0.1036824 0.4943879
ranef(model_nested)
$`mountainRange:site`
(Intercept)
Bavarian:a -2.46727186
Bavarian:b -3.05278205
Bavarian:c 3.73563393
Central:a -6.38640673
Central:b 0.84540907
Central:c 6.17300851
Emmental:a -3.02659531
Emmental:b 2.29768424
Emmental:c -0.03273939
Julian:a 2.42006432
Julian:b 2.20653961
Julian:c -3.44048746
Ligurian:a 0.99876698
Ligurian:b -0.84624351
Ligurian:c -0.80816294
Maritime:a -3.67613987
Maritime:b -0.63034574
Maritime:c 5.87130429
Sarntal:a 3.65403344
Sarntal:b -4.91065013
Sarntal:c 2.27894013
Southern:a -0.68072360
Southern:b 2.18570243
Southern:c -2.70853837
$mountainRange
(Intercept)
Bavarian -25.313507
Central 8.965609
Emmental -10.804656
Julian 16.826066
Ligurian -9.300801
Maritime 22.198277
Sarntal 14.502523
Southern -17.073511La prédiction se fait en additionnant les différentes composantes.
# Ordonné à l'origine
coefficients(summary(model_nested))[1, 1] +
# Taille des 6 premiers dragons * coefficient de cette variable
dragons$bodyLength[1:6] * coefficients(summary(model_nested))[2, 1] +
# Coefficient dû à la montagne (Bavarian pour les 6 premiers)
ranef(model_nested)$mountainRange["Bavarian", ] +
# Coefficient dû au site dans la montagne (Bavarian:a pour les 6 premiers)
ranef(model_nested)$`mountainRange:site`["Bavarian:a", ]
[1] 20.77181 20.87488 20.78896 20.88135 20.99793 20.93278Et on retrouve le même résultat
augment(model_nested, dragons)$.fitted[1:6]
[1] 20.77181 20.87488 20.78896 20.88135 20.99793 20.93278Probabilités critiques
Si veut les probabilités critiques pour chaque variable, on a besoin de charger le package lmerTest.
library(lmerTest)
model_mixed <- lmer(testScore ~ bodyLength + (1|mountainRange),
data = dragons)effect | group | term | estimate | std.error | statistic | df | p.value |
fixed | (Intercept) | 43.709 | 17.135 | 2.551 | 177.466 | 0.012 | |
fixed | bodyLength | 0.033 | 0.079 | 0.422 | 472.667 | 0.673 | |
ran_pars | mountainRange | sd__(Intercept) | 18.430 | ||||
ran_pars | Residual | sd__Observation | 14.960 |
sigma | logLik | AIC | BIC | REMLcrit | df.residual |
14.960 | -1995.601 | 3999.203 | 4015.898 | 3991.203 | 476 |
La variable bodyLength n’est pas significative.
Conclusion
On a un joli paradoxe de Simpson :)